Disease-Gene Association
1. Introduction
Extraction of relations between specific diseases and their relevant genes or proteins is an important task in bioinformatics. Our goal is to automatically extract such relations by mining the literature containing disease-gene relations. Considering the utility of the results of this approach, we identified disease and gene names that can be mapped to ID tags of public biomedical databases. Moreover, considering that genetics experts will use our results, we classified them based on topics that show the types of disease-gene relations.
To map recognized gene and protein names to ID tags, we used a dictionary matching approach. However, the dictionary matching approach usually yields too many false positives. In order to reduce the number of false positives, we filter the results using a machine learning method. We developed a Maximum Entropy (ME)-based disease and gene name recognizer, a relation extractor and a topic-classified relation extractor, and used them in a corpus-based approach.
The development corpus is a subset of MEDLINE abstracts dealing with prostate cancer and gastric cancer. It was annotated with disease and gene relations, based on two topics: “etiology” and “clinical marker”. To extract relations based on these topics, we collected various features including aliases, synonyms, acronyms and full names of candidate disease and gene names. These features were obtained from abstracts, using context extension and coreference recognition. We used them to train the Maximum Entropy (ME)-based disease and gene name recognizer, relation extractor, and topic-classified relation extractor.
2. Contents
Figure 1 describes an example of topic-classified relations. The output of our system is the list of relevant gene names with respect to a certain disease based on a topic, and their evidential sentences.
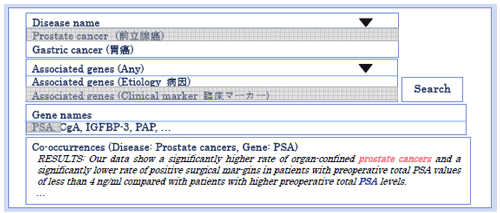 Figure 1: An example of topic-classified relations
Figure 1: An example of topic-classified relationsOur system first collects sentences that contain at least one pair of disease and gene names, using the dictionary-based longest matching technique. We used machine learning-based methods to filter out false positives from the dictionary matching results. The named entity recognizer filtered out a lot of false positive disease and gene names, the relation extractor filtered out a lot of false positive relations, and the topic-classified relation extractor filtered out a lot of false positive topic-classified relations (Figure 2).
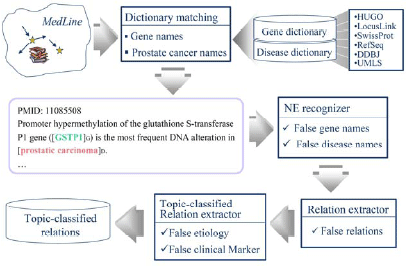 Figure 2: System architecture (Extraction of Disease-Gene Association)
Figure 2: System architecture (Extraction of Disease-Gene Association)3. Dictionary matching and annotation
We constructed disease and gene dictionaries and used them to augment the corpus of sentences collected in step one (“disease-gene-pairs sentences”). Thus, all disease and gene names in the results of the proposed method have ID tags that are used in the publicly available biological databases (HUGO, LocusLink, SwissProt, RefSeq, DDBJ and UMLS). When a disease-gene pairs sentence contained more than one disease name and more than one gene name, the system made sufficient copies of the sentence to accommodate all possible disease-gene name pairs. We call these copies disease-gene pair instance sentences, which are the input units of our relation extraction system. Each instance sentence is a candidate for a topic-classified relation between one prostate cancer and one gene. We randomly chose 2,999 and 1,000 instance sentences for prostate cancer and gastric cancer, respectively. These instance sentences were topic-annotated by six biologists. This corpus was used to train the named entity filter, the relation filter, and the topic-classified relation filter.
4. Named entity recognition
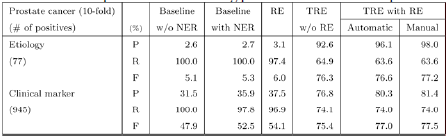
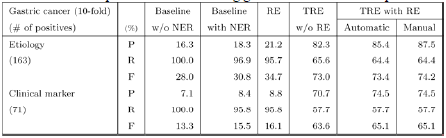
We used a machine learning-based named entity recognition method to filter out numerous false positive disease and gene names from the dictionary matching results. Maximum Entropy approaches have been developed and used to train the named entity filter, considering orthographic features and syntactic features. To obtain syntactic features, we used the ENJU full parser[1] and the GENIA Part-of-Speech Tagger[2]. Named entity recognition improved the precision of topic-classified relation extraction at the cost of a small reduction in relative recall compared to experiments using the dictionary matching method. (See Table 1 and Table 2).
5. Relation extraction
Disease and gene name pairs co-occurring in a sentence have many potential relations. However, these co-occurring pairs also have numerous false-positive relations. We developed a Maximum Entropy (ME)-based relation extractor to filter out false positive relations. Thus, candidate relations which remain after the relation filtering can be classified as etiology or clinical marker.
6. Topic-classified relation extraction
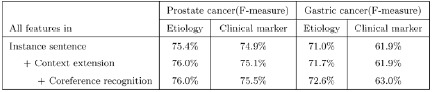
Our relation extraction method showed 95.5% and 89.5% F-score for extracting prostate cancer-gene relations and gastric cancer-gene relations, respectively. However, this relation extraction method showed very low performance for extracting topic-classified relations (See Table 1 and Table 2). This observation motivates development of the topic-classified relation extraction method. A topic-classified relation is a semantic relation, and the data sparseness problem should be considered. To reduce the influence of the data sparseness problem, we employed coreference recognition and context extension. We considered full names, acronyms, aliases, and synonyms of candidate disease and gene names for coreference recognition, because theses various expressions of biomedical names are frequently used in the literature. We dealt with abstracts to extend the context. In context extension, we exploited a powerful property of human language: The context (including word sense) of a target word is highly consistent within any given document [3]. We expected that there would be more information in full abstracts than in instance sentences.
7. Experimental results and Discussion
Table 1 and Table 2 describe the performance of the topic-classified relation extraction using all methods. Numbers in the first column represent the number of positive instance sentences for each topic. To evaluate the systems in this section, we performed 10-fold cross validation using 2,999 prostate cancer- and 1,000 gastric cancer-related instance sentences. Experiments with the title of Baseline without NER (named entity recognition) used only dictionary matching results, experiments with the title of Baseline with NER used both dictionary matching results and disease and gene name recognition results, and experiments with the title of RE used a relation extractor which included the dictionary matching and disease and gene name recognition results.
In the next experiments, we used a Maximum Entropy-based machine learning technique for extraction of disease and gene relations based on the topics of etiology and clinical marker. In experiments with the title of TRE without RE, we applied Maximum Entropy-based topic-classified relation extraction, but did not use a relation filtering method. The input data for these experiments were the results of dictionary matching and disease- and gene-name recognition. Experiments with the title of TRE with RE used all methods in our system: dictionary matching, disease and gene name recognition, relation extraction, and topic-classified relation extraction.
We also compared the performance of two types of experiments: automatic and manual. In experiments with the title of automatic, the machine learning-based topic-classified relation extractor used the results of machine learning-based systems for disease and gene name recognition and relation extraction. In experiments with the title of manual, the relation extractor was applied to human-annotated disease names, gene names, and relations.
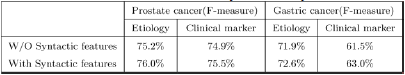
Table 3 describes the effectiveness of context extension and coreference recognition. The topic-classified relation extraction method using context extension and coreference recognition obtained better performance (F-score). Table 4 shows the effectiveness of syntactic features for topic-classified relation extraction. Using the syntactic features, we achieved a better score. In Table 5, we compared one-phase TRE and Two-phase TRE.

In two-phase TRE, we conducted TRE after filtering by RE. Two-phase TRE shows better performance.
8. Conclusion and Future work
A series of experimental results revealed four important findings: (1) A carefully designed named entity filtering and relation filtering method can improve the performance of topic-classified relation extraction. (2) Features that were obtained by context extension and coreference recognition improved the performance of topic-classified relation extraction. (3) Syntactic features improved the performance of topic-classified relation extraction. (4) The Maximum Entropy-based topic-classified relation extraction approach achieved encouraging results for both prostate cancer and gastric cancer.
The approach described here is heavily dependent upon manually constructed resources. To reduce the cost to build dictionary and annotated corpus, we plan to employ some techniques such as active learning [4] and domain adaptation [5]. The proposed approach focuses on reducing false positives of dictionary matching technique. To deal with false negatives of dictionary matching technique, we plan to employ some techniques such as spelling variation [6].
References
- Yusuke Miyao and Jun'ichi Tsujii, Probabilistic Disambiguation Models for Wide-Coverage HPSG Parsing., Proceedings of the 43rd Annual Meeting of the Association for Computational Linguistics (ACL), 2005: pp. 83-90.
- GENIA Part-of-Speech Tagger v0.3, http://www-tsujii.is.s.u-tokyo.ac.jp/GENIA/postagger/, 2004.
- David Yarowsky, (1995), Unsupervised Word Sense Disambiguation Rivaling Supervised Methods. Proceedings of the 33rd Annual Meeting of the Association for Computational Linguistics (ACL), pp. 189-196.
- Charles C. Bonwell and James A Eison, Active Learning: Creating Excitement in the Classroom., AEHE-ERIC Higher Education Report No.1, 1991: Washington, D.C.: Jossey-Bass. ISBN 1-87838-00-87.
- Hal Daume III and Daniel Marcu, Domain Adaptation for Statistical Classifiers. International Journal of Artificial Intelligence Research (JAIR)., 2006: Vol. 26, pp. 101-126.
- Yoshimasa Tsuruoka and Jun'ichi Tsujii, Boosting Precision and Recall of Dictionary-Based Protein Name Recognition., Proceedings of the ACL-03 Workshop on Natural Language Processing in Biomedicine., 2003: pp. 41-48.
