Event Annotation and Recognition
1. Introduction
In this work we have framed the event recognition problem as a binary classification problem, determining whether a given sentence mentions a specific type of event or not. This report describes our work on recognizing the 'cellular physiological process (GO:0050875)' type of events. However, the method described here can be easily generalized to other types of events, as long as there is a corpus annotated for the desired event type.
To tackle the sentence classification problem, we made use of three major resources. The Maximum Entropy Classifier modeling toolkit was used to automatically train classifiers. The GENIA event annotated corpus was used to provide the toolkit with training material. The Enju syntactic parser was used to gain a better handle on natural language expressions.
2. GENIA event annotated corpus
The GENIA event annotated corpus is a collection of Medline abstracts with biological event annotation. In this corpus, all the abstracts were read by human annotators. Biological events recognized by the annotators were annotated, using a predefined set of descriptors which correspond to classes in Gene Ontology. The corpus contains 958 abstracts, annotated for 37 classes of events.
In this context, “EVENT” is defined as a process involving change of certain states of biological entities. Examples from text which do not involve such “status changing” are not annotated as events, even if they are important biologically.
Examples
- Protein expression (IL2 gene expression)
- Initiation of transcription (initiation of IL2 transcription)
- Receptor activation (TCR triggering)
- Promotion or inhibition of cell proliferation (T cell proliferation is initiated by …)
- Formation of the complex (CD229-Grb2 complex formation)
- Enzymatic reaction (PT catalyzed ADP-ribosylation)
- Virus infection (HIV infection)
In the annotated corpus, a sentence is followed by one or more EVENT elements, each of which indicates an event mentioned in the sentence.
- EVENT: EVENT is the term(s) that mean(s) biological status changing. EVENT is described by TYPE, THEME, CAUSE and CLUE.
- TYPE: TYPEs of the event are the categories of the EVENT, chosen from the predefined set of event classes, the "EVENT ontology".
- THEME: THEMEs are the targets of the EVENT, designated by an “ID”, term, event or sentence, etc.
- CAUSE: CAUSEs are the causes of the EVENT, designated by an “ID”, term, event or sentence, etc.
- CLUE: CLUE elements are the places where location information can be recorded. The information is classified and the tagged in the sentence. Tags show the object, cause etc. of the EVENT in the sentence. The rules for tagging are described in Section 3.5.
- COMMENT: COMMENT is a free description for the EVENT.
Figure 1 shows the hierarchy of the GENIA event ontology. The numbers after the concepts indicate the frequency of the appropriate annotations in the corpus.
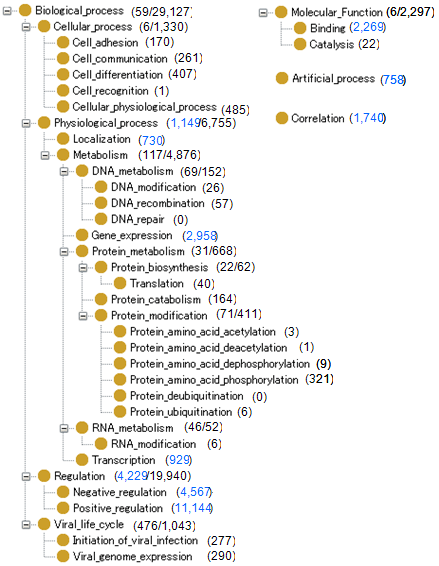 Figure 1: GENIA event ontology
Figure 1: GENIA event ontologyFigure 2 shows an example of event annotation. The text appearing in the black box is the sentence annotated with biological terms, and each term is assigned a termID.
For an identified event in the given sentence,
- classify the type of events and record the text span giving the clue of it (ClueType).
- identify the theme of the events and record the text span linking the theme to the event (LinkTheme).
- identify the cause of the events and record the text span linking the cause to the event (LinkCause).
- record the environment (location, time) of the events (ClueLoc, ClueTime).
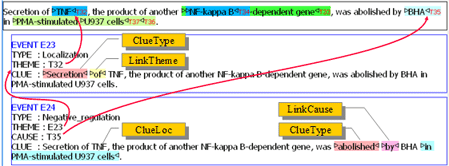 Figure 2: Example of event annotation
Figure 2: Example of event annotationOut of 7,992 annotated sentences, there are 337 sentences that have been annotated with 'cellular physiological process' type of events. A simple baseline method for identifying sentences that contain mentions of 'cellular physiological process’ could classify all sentences as positive. In this baseline method, the recall is 100% (the upper bound) while precision is 4.2% (the lower bound).
3. Experiments
In accordance with the problem definition, a number of binary classifiers were induced from the GENIA event-annotated corpus. When we work with machine learning methods, a training example is usually represented as a set of features. Different classifiers are induced depending on the feature representations. To handle the problem of sentence classification, we experimented with several representations of sentence.
2.3.1. Bag-of-words classifier
A Bag-of-words is a common representation of natural language text. In this representation, a sentence is treated as just a bag of words without considering the order of words. This representation is easy to generate and known to work reasonably well. A standard practice often involves stemming each word and removing stop words. From this representation of the GENIA corpus we induced a maximum entropy classifier, which showed performance of 40.3% recall and 43.8% precision.
2.3.2. Clueword classifier
The GENIA event annotation includes clue expressions which are supposed to give a clue to the determination of an event type. Figure 3 shows an example of GENIA event annotation. The example sentence is annotated with an event description. The type of event is determined as a cellular physiological process due to the word “proliferation”, which is recorded as the clue expression (for the type of event).
The effects of prostaglandin E2 (PGE2) on cytokine production and proliferation of the CD4+ human helper T cell clone SP-B21 were investigated. |
|
EV1 |
TYPE: Cellular_physiological_process |
From the clue expressions collected from the GENIA annotation corpus, we generated a list of keywords for the recognition of cellular physiological process. Figure 4 shows some of the stemmed keywords, with statistics. For example, the keyword stem ‘proliferat’ appears 153 times in total and 109 times with annotation in the corpus. We can thus expect that if we collect sentences with the word ‘proliferat’, 71% of them may be mentioning a cellular physiological process.
|
keyword |
precision |
# appearance |
# annotation |
|---|---|---|---|---|
1 |
diapedesis |
100% |
3 |
3 |
2 |
cytolysis |
100% |
2 |
2 |
3 |
division |
100% |
2 |
2 |
4 |
rolling |
100% |
2 |
2 |
5 |
mitogenesis |
85% |
7 |
6 |
6 |
cytoly |
8% |
10 |
8 |
7 |
expansion |
75% |
4 |
3 |
8 |
lysis |
72% |
11 |
8 |
9 |
syncyti |
71% |
7 |
5 |
10 |
proliferat |
71% |
153 |
109 |
… |
||||
42 |
divid |
8% |
65 |
5 |
43 |
heterologous |
6% |
31 |
2 |
44 |
clear |
5% |
40 |
2 |
45 |
stimulat |
4% |
519 |
23 |
46 |
form |
4% |
222 |
8 |
Note that the list also includes keywords of low precision. For example, if we pick a sentence with the keyword ‘divid’, it may be mentioning a cellular physiological process with a probability of 8%. Overall, if we take sentences with these keywords to be sentences mentioning a cellular physiological process, the recall might be 94.6% and the precision 17.2%.
We trained a maximum entropy classifier to raise the precision. This time we used a feature representation of sentences that included the keywords and the surrounding words as features. With this classifier making use of keyword features, we achieved a performance of 58.5% of recall and 64.7% of precision.
2.3.3. Clueword classifier with syntactic patterns
In the previous section, we describe a classifier making use of words that surround keywords to disambiguate the keywords. In this section we describe a classifier making use of syntactic patterns instead of just surrounding words.
We parsed all sentences in the GENIA event-annotated corpus using Enju, a syntactic parser, to get access to the syntactic structure of the sentences. An analysis of the parsed corpus indicates that there are prevailing syntactic patterns with which cellular physiological processes are often mentioned. Figure 5 lists the three patterns. Pattern 1 describes the case where an entity which is a theme of an event appears in the same noun phrase as the keyword representing the event. Pattern 2 describes the case where the theme-entity appears in a noun phrase which is connected to the noun phrase including the keyword by the preposition ‘of’. Pattern 3 describes the case where the theme-entity appears in a noun phrase which is a semantic argument of the verb phrase including the keyword.
| Pattern1 | [KEYWORD THEME_ENTITY]NP |
|---|---|
| Examples | highly polarized TH2 clone, cell proliferation |
| Pattern2 | [KEYWORD]NP of [THEME_ENTITY]NP |
| Examples | proliferation of cell, grow of multiple mold-4 cem cell |
| Pattern3 | [KEYWORD]VP·(ARG)·[THEME_ENTITY]NP |
| Examples | proliferate human monocyte, kill fibroblastic keratinocyte-derived human cell |
Based on the patterns, we developed a feature representation of sentences which is designed to reflect the three syntactic patterns. A maximum entropy classifier induced from this representation of the GENIA corpus showed a performance of 61.5% recall and 65.2% precision.
4. Future directions
There are several directions for improvement.
- Richer global features: The current experiments use the event-annotated GENIA corpus. The recall and precision are calculated by 10-fold cross-validation. However, the GENIA corpus is skewed as a corpus for representing all of Medline. We may have to use more global features which capture text classes of abstracts.
- Event classes: We assume that the current set of event classes gives us a reasonable level of abstraction for sentence classification. However, we may have to use lower or upper classes from the Gene Ontology (GO) to have better classifiers, even though this requires re-annotation.
In any case, it is important to produce a set of classifiers for all event classes in order to claim that our method is robust and scalable enough for actual application.
References
- Tateisi, Yuka and Jun'ichi Tsujii. Part-of-Speech Annotation of Biology Research Abstracts. In the Proceedings of 4th International Conference on Language Resource and Evaluation (LREC2004). IV. Lisbon, Portugal, pp. 1267-1270, May 2004.
- Tateisi, Yuka, Akane Yakushiji, Tomoko Ohta and Jun’ichi Tsujii. Syntax Annotation for the GENIA corpus. In the Proceedings of the IJCNLP 2005, Companion volume. Jeju Island, Korea, pp. 222--227, October 2005.
- Kim, Jin-Dong, Tomoko Ohta, Yuka Teteisi and Jun'ichi Tsujii. GENIA corpus - a semantically annotated corpus for bio-textmining. Bioinformatics. 19(suppl. 1). pp. i180-i182, Oxford University Press, 2003. ISSN 1367-4803.
- Miyao, Yusuke and Jun'ichi Tsujii. Probabilistic disambiguation models for wide-coverage HPSG parsing. In the Proceedings of ACL 2005. Ann Arbor, Michigan, pp. 83-90, June 2005.
