MEDIE — Retrieval of Relational Concepts using Semantic Structures
1. Introduction
Rapid expansion of text information has motivated the development of efficient methods of accessing information in huge texts. For example, biomedical researchers deal with a massive quantity of publications; MEDLINE contains approximately 15 million references to journal articles in life sciences, and its size is rapidly increasing, at a rate of more than 10% yearly. Researchers would like to be able to search this huge textbase for relational concepts such as protein-protein or gene-disease associations. However, keyword-based IR techniques are insufficient to describe such relations precisely.
This section describes MEDIE (Figure 1), a system for the accurate real-time retrieval of relational concepts from MEDLINE (Miyao et al. 2006). This system consists of the off-line processing to compute semantic structures and the on-line processing to search for semantic structures that match a user’s query (Figure 2).
- Off-line processing: An HPSG parser (Miyao et al. 2005) and a term recognizer (Tsuruoka et al. 2004) are applied in order to create MEDLINE annotated with predicate argument structures and identifiers in ontology databases.
- On-line processing: User input is converted into queries of the extended region algebra (Masuda et al. 2003). A search engine retrieves sentences having semantic annotations that match the queries.
The accurate retrieval of relational concepts is attained because we can precisely describe relational concepts using semantic annotations. In addition, real-time retrieval is possible because semantic annotations are computed in advance.
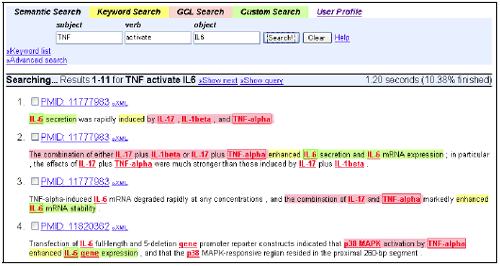 Figure 1:MEDIE
Figure 1:MEDIE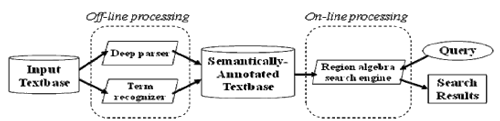 Figure 2:The architecture of MEDIE
Figure 2:The architecture of MEDIE2. HPSG parsing
HPSG is a contemporary syntactic theory that explains a mapping between a sentence and its meaning. HPSG parsing can therefore output not only phrase structures but also predicate argument structures, which are canonicalized representations of sentence meanings and express semantic relations of words explicitly. In this research, we applied an accurate and efficient HPSG parser (Miyao et al. 2005) to MEDLINE. We used two geographically separated computer clusters having 170 nodes (340 Xeon CPUs) by using GXP (Taura 2004), and finished parsing the entire MEDLINE in nine days (Ninomiya et al. 2006).
| Symbol | CRP |
|---|---|
Name |
C-reactive protein, pentraxin-related |
Species |
Homo sapiens |
Synonym |
MGC88244, PTX1 |
3. Term recognition
Ontology databases are collections of words and phrases in specific domains. Such databases have been constructed extensively for the systematic management of domain knowledge by organizing textual expressions of ontological entities. For example, Table 1 shows an entry of GENA, a database of genes and proteins. “Symbol”, “Name”, and “Species” denote short forms, nomenclatures of genes, organism species, respectively. “Synonym” is a list of synonyms. Table 1 indicates that CRP, MGC88244, and PTX1 denote the same gene. Hence, these resources enable us to canonicalize variations of textual expressions of ontological entities. We applied a dictionary-based term recognition algorithm (Tsuruoka et al. 2004) in order to annotate genes, proteins and diseases with their ontological identifiers.
Verbal expressions are also important for the description of relational concepts. For example, activation of proteins may be expressed by “activate” or “enhance”. Although the number of such expressions is much smaller than that of technical terms, an event ontology was not available. We therefore developed an ontology from scratch. We investigated 500 abstracts extracted from MEDLINE, and classified 167 frequent expressions into 18 event types. These expressions in MEDLINE were automatically annotated with event types.
4. Extended Region Algebra
In this research, we adopted extended region algebra that allows us to search structured texts with tags with nested and cross boundaries. Extended region algebra is defined as a set of operators on regions, i.e., word sequences (Table 2). A and B denote regions, and results of operations are also regions. Four containment operators (>, >>, <, <<) represent ancestor/descendant relations in XML. For example, “A > B” indicates that A is an ancestor of B. In search algorithms for region algebra, the cost of retrieving the first answer is constant, and that of an exhaustive search is bounded by the lowest frequency of a word in a query.
| [tag] | Region covered with “<tag>” |
|---|---|
A > B |
A containing B |
A >> B |
A containing B (A is not nested) |
A < B |
A contained by B |
A << B |
A contained by B (B is not nested) |
A & B |
A and B |
A | B |
A or B |
For example, a query to search for “CRP excludes something” is expressed as:
[sentence] >>(([word arg1="$subject"] > exclude) & ([phrase id="$subject"] > CRP))
This formula indicates that a sentence contains “exclude”, and its first argument (“arg1”) phrase includes “CRP”. A predicate argument relation is expressed by the variable, “$subject”. Since it is difficult for users to input such queries directly, this system automatically creates a query of extended region algebra given subject x, object y, and verb v by replacing x, y, and z with ontological identifiers when possible.
| # papers | 14,785,094 |
# abstracts |
7,291,857 |
# sentences |
70,935,630 |
# words |
1,462,626,934 |
# successfully parsed sentences |
69,243,788 |
# predicate argument relations |
3,094,105,383 |
# terms (genes) |
84,998,621 |
# terms (proteins) |
27,471,488 |
# terms (diseases) |
19,150,984 |
# terms (event expressions) |
51,810,047 |
Size of the original MEDLINE |
9.3 GByte |
Size of the semantic annotations |
292 GByte |
Size of the index files |
954 GByte |
| 1 | something inhibit ERK2 |
2 |
something trigger diabetes |
3 |
adiponectin increase something |
4 |
TNF activate IL6 |
5 |
dystrophin cause disease |
6 |
macrophage induce something |
7 |
something suppress MAP phosphorylation |
8 |
something enhance p53 (negative) |
5. Experiments
The data used in the experiments consists of all articles registered in MEDLINE at the end of 2004. Table 3 shows the size of data before/after the off-line processing. Table 4 is a list of queries used in the following experiments. “something” and “disease” indicate that any word and disease name can appear, respectively, while “(negative)” indicates negative mentions. These were selected by a biologist, and express typical relational concepts that a biologist may wish to find.
Table 5 summarizes the results of the experiments. We have two parameters to vary: whether to use predicate argument structures and whether to use ontological identifiers. “Keyword search” is based on occurrences of keywords, while “semantic search” uses predicate argument relations. The postfixes of query numbers denote whether ontological identifiers are used. X-1 used no ontologies, X-2 used only the term ontology, X-3 used both the term and event expression ontologies. Accuracy is reported in precision and in recall relative to the results of keyword search. At most 100 sentences were retrieved for each query, and the results were merged and shuffled. A biologist judged the shuffled sentences (1,839 sentences in total).
The results show that the semantic search exhibited impressive improvements in precision. The precision was over 80% for most queries and was nearly 100% for Queries 4 and 5. This indicates that predicate argument structures are effective for representing relational concepts precisely. The increase of the number of retrieved results indicates the effectiveness of ontologies. In particular, the results show that the event expression ontology was indispensable.
Query |
Keyword search |
Semantic search |
|||||
|---|---|---|---|---|---|---|---|
# ans. |
time |
preci- |
# ans. |
time |
preci- |
relative |
|
1-1 |
252 |
0.00/1.5 |
74% |
143 |
0.01/2.5 |
96% |
69% |
1-2 |
348 |
0.00/1.9 |
61% |
174 |
0.01/3.1 |
89% |
69% |
1-3 |
884 |
0.00/3.2 |
50% |
292 |
0.01/5.3 |
91% |
42% |
2-1 |
125 |
0.00/1.8 |
45% |
27 |
0.02 |
85% |
38% |
2-2 |
113 |
0.00/2.9 |
40% |
26 |
0.06/4.0 |
85% |
48% |
2-3 |
6529 |
0.00/12.1 |
42% |
662 |
0.01/1527 |
76% |
19% |
3-1 |
287 |
0.00/1.5 |
20% |
30 |
0.05/2.4 |
80% |
30% |
3-2 |
309 |
0.01/2.1 |
21% |
32 |
0.10/3.5 |
81% |
29% |
3-3 |
338 |
0.01/2.2 |
24% |
39 |
0.05/3.6 |
82% |
33% |
4-1 |
4 |
0.26/1.5 |
0% |
0 |
2.44/2.4 |
- |
- |
4-2 |
195 |
0.01/2.5 |
9% |
6 |
0.09/4.1 |
83% |
22% |
4-3 |
2063 |
0.00/7.5 |
5% |
94 |
0.02/10.5 |
95% |
40% |
5-2 |
287 |
0.08/6.3 |
73% |
116 |
0.05/14.7 |
97% |
51% |
5-3 |
602 |
0.01/15.9 |
50% |
122 |
0.05/14.2 |
96% |
46% |
6-1 |
10698 |
0.00/42.8 |
14% |
1559 |
0.01/3015 |
65% |
71% |
6-3 |
42106 |
0.00/3380 |
11% |
2776 |
0.01/5100 |
61% |
45% |
7 |
87 |
0.04/2.7 |
39% |
15 |
0.05/4.2 |
67% |
29% |
8 |
1812 |
0.01/7.6 |
19% |
84 |
0.20/29.2 |
87% |
37% |
6. Conclusion and Future Work
We demonstrated a text retrieval system for MEDLINE that exploits pre-computed semantic annotations. Experiments revealed that the proposed system is sufficiently efficient for real-time retrieval and that the precision of retrieval was remarkably high. While this system focused on subject-verb-object relations, semantic structures include other semantic relations and they may be exploited to search for various concepts such as experimental settings and modality. Although the work described here focused on MEDLINE, the NLP tools used in this system are domain/task independent and this framework will be applicable to other domains, such as patent documents.
Reference
- Miyao, Yusuke, Tomoko Ohta, Katsuya Masuda, Yoshimasa Tsuruoka, Kazuhiro Yoshida, Takashi Ninomiya and Jun'ichi Tsujii. Semantic Retrieval for the Accurate Identification of Relational Concepts in Massive Textbases. In the Proceedings of COLING-ACL 2006. Sydney, Australia, pp. 1017-1024, July 2006.
- Miyao, Yusuke and Jun'ichi Tsujii. Probabilistic disambiguation models for wide-coverage HPSG parsing. In the Proceedings of ACL 2005. Ann Arbor, Michigan, pp. 83-90, June 2005.
- Tsuruoka, Yoshimasa and Jun'ichi Tsujii. Improving the Performance of Dictionary-based Approaches in Protein Name Recognition. Journal of Biomedical Informatics. 37(6). pp. 461-470, Elsevier, 2004.
- Masuda, Katsuya, Takashi Ninomiya, Yusuke Miyao, Tomoko Ohta and Jun'ichi Tsujii. A Robust Retrieval Engine for Proximal and Structural Search. In the Proceedings of HLT-NAACL 2003 Short papers. Edmonton, Canada, pp. 58-60, May 2003.
- Taura, Kenjiro. GXP: An interactive shell for the grid environment. In the Proceedings of IWIA2004. 2004.
- Ninomiya, Takashi, Yoshimasa Tsuruoka, Yusuke Miyao, Kenjiro Taura and Jun'ichi Tsujii. Fast and Scalable HPSG Parsing. Traitement automatique des langues (TAL). 46(2). Association pour le Traitement Automatique des Langues, 2006.
