CHEBI corpus
Description
The ChEBI corpus contains 199 annotated abstracts and 100 annotated full papers. All documents in the corpus have been annotated for named entities and relations between these. In total, our corpus provides over 15000 named entity annotations and over 6,000 relations between entities. The entities were annotated according to the requirements of the curators of the ChEBI database. We primarily annotated mentions of metabolites, as well as other entities that were capable of bearing interesting information related to metabolites, such as: Chemicals, Proteins, Species, Biological Activity and Spectral Data. The entities were annotated with an inter-annotator agreement of 0.80-0.89 (F1 score, strict-matching). The relations provided further information about the links between metabolites and other entities. We chose the following categories: Isolated From, Associated With, Binds With, Metabolite Of. The ChEBI corpus can be used to investigate interesting lexical properties of metabolites and related entities. In addition, it can be used to train machine learning algorithms to recognise the entities and relations that have been annotated.
Availability
The corpus is available for download.
Please observe the terms of the licence below in any research arising from this corpus.
Licence

The annotations in the CHEBI corpus are licensed under a Creative Commons Attribution 4.0 International License.
PLEASE CITE THE FOLLOWING PAPER IN ANY RESEARCH ARISING FROM THIS CORPUS:
Shardlow, M., Nguyen, NTH., Owen, G., Turner, S., O'Donovan, C., Leach, A., McNaught, J., and S. Ananiadou (2018) A New Corpus to Support Text Mining for the Curation of Metabolites in the ChEBI Database. In Proceedings of LREC, Miyazaki, Japan, pp. 280-285.
Motivation
The ChEBI (Chemical Entities of Biological Interest) database is a freely available, electronic dictionary and ontology of small molecules. ChEBI was created to help researchers in the field of molecular biology who need to know the structure, names, and properties of the small molecules that they encounter in their research. There are a number of freely-available chemical databases. Most of them are created by an automatic 'pipeline' process and contain information on polymers, industrial chemicals, synthetic intermediates, etc. Their sheer size creates problems for users, as any search may result in hundreds or even thousands of answers. For non-expert users it is very difficult to determine which, if any, is the compound they are really looking for. By contrast, the focus of the ChEBI database is on high quality rather than quantity. ChEBI is manually curated and focuses on the requirements of the molecular biology community. Manual curation assures the high quality of the database, but it also makes it an expensive database to produce, particularly so for our focus of metabolites. Users who would like to add a new metabolite, may only know a research code or a trivial name from which it is not possible to deduce a structure. The curator then has to search through scientific literature to find as much information as possible about the new metabolite. The information is likely to include:
- In which species is it present?
- Does it have any interesting biological properties, applications, etc? (E.g., Biological activity)
- Is there any spectral data available that indicates the structure?
- From which chemical's metabolism does it derive?
Annotation Scheme
We annotated the following entities:
Metabolite: A chemical which has been produced by, detected in, or isolated from a living organism, where this is clear from the context of the paper (e.g. Nitrosobenzene, 11-Deoxycorticosterone, sclerotiorin).
Chemical: Any name that is used to define 'small' chemicals (those that are not proteins, nucleic acids, etc.). Includes molecules, salts, class names (e.g. benzoate esters; indole alkaloids; etc.) and groups (parts of molecules) - e.g. methyl group; benzyl substituent; alanine residue. . . )
Protein: Any protein or large polypeptide (usually one that is too big to be drawn by normal chemical drawing software). All enzymes and receptors are considered to be proteins in our scheme (e.g. 4-Dihydroxyphenylalanine decarboxylase, Dopa decarboxylase).
Species: Any entity referring to a formal name for a living organism or from which the name can be inferred (e.g. 'volunteer', 'patient' implies 'human').
Biological Activity: An effect/consequence that a chemical entity has on a biological system. Examples may include affecting the activity (e.g. by inhibition or activation) of a particular enzyme; growth regulator; antimicrobial agent; apoptosis inducer; antiinflammatory; flavour enhancer; etc.
Spectral Data: data arising from spectrometry. e.g., 1H-NMR, 13C-NMR, MS, X-ray, IR, etc. Where two or more spectroscopic techniques are present, each should be tagged separately.
And the following relations:
Isolated_From(Metabolite, Species): A metabolite was isolated from or detected in a specific species.
Associated_With(Chemical / Metabolite, Biological Activity / Spectral Data): A chemical or metabolite is linked to a particular biological activity or spectral data.
Binds_With(Chemical / Metabolite, Protein): A chemical or metabolite interacts with (e.g.binds) and affects the behaviour of a biological target.
Metabolite of(Metabolite, Chemical): A metabolite is derived from the metabolism of a related compound.
An example document annotated according to this scheme is shown in Fig 1.
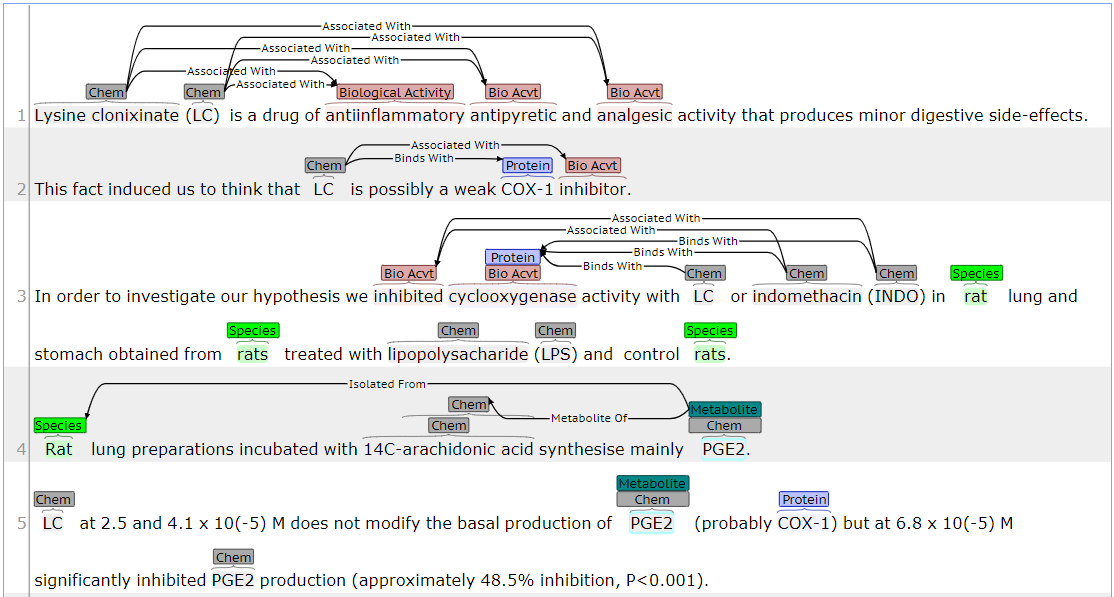 Fig 1. - ChEBI corpus annotation example.
Fig 1. - ChEBI corpus annotation example.
Evaluation
We calculate strict and relaxed matching between 2 annotators on the set of abstracts (200 texts). We show an average agreement of strict:0.847/relaxed:0.930 for entities and an average agreement of strict:0.623/relaxed:0.773 for relations. All agreement is reported as F1 scores.
Acknowledgements
This work was funded by the OpenMinTeD project EC/H2020/654021.
Featured News
- 1st Workshop on Misinformation Detection in the Era of LLMs - Presentation slides now available
- Prof. Ananiadou appointed Deputy Director of the Christabel Pankhurst Institute
- ELLIS Workshop on Misinformation Detection - Presentation slides now available
- Prof. Sophia Ananiadou accepted as an ELLIS fellow
- BioNLP 2025 and Shared Tasks accepted for co-location at ACL 2025
- Prof. Junichi Tsujii honoured as Person of Cultural Merit in Japan
Other News & Events
- AI for Research: How Can AI Disrupt the Research Process?
- CL4Health @ NAACL 2025 - Extended submission deadline - 04/02/2025
- Invited talk at the 15th Marbach Castle Drug-Drug Interaction Workshop
- Participation in panel at Cyber Greece 2024 Conference, Athens
- Shared Task on Financial Misinformation Detection at FinNLP-FNP-LLMFinLegal








