KISTI Pathway project

Background
The construction of detailed, machine-readable models of biomolecular pathways is a major goal of systems biology, and hundreds of models capturing the physical entities and reactions involved in various pathways are already available from repositories such as the BioModels Database and the PANTHER Pathway repository.
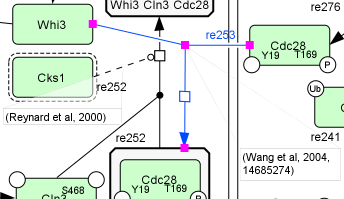
However, the manual construction, quality control and maintainance of pathway models is a demanding and expensive effort, and one of the key challenges in this effort is the information overload caused by the exponential growth of the biomedical scientific literature: currently, a new citation is added into the PubMed literature database on average once every 40 seconds.
Biomedical text mining systems are increasingly capable of creating rich structured representations of information automatically extracted from literature. Such text mining systems open many opportunities for supporting the curation, validation, and updating of pathway models.
Project
Following the joint signing of a memorandum of understanding, NaCTeM is collaborating with the Korea Institute of Science and Technology Information (KISTI) to develop the next generation of information extraction and text mining systems for supporting and automating various aspects of biomolecular pathway model curation.
Building on the PathText text mining integration technology for pathways, text mining systems such as MEDIE, event extraction tools such as EventMine, we are developing methods for identifying literature relevant to specific reactions in pathway models and for automatically analysing documents to extract event structures that capture the full semantics of pathway reactions.
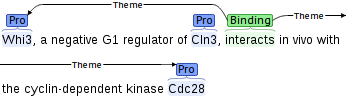
Key among the aims of the project are the development of advanced ranking technology for determining the relevance of documents to given pathway reactions and the extension of the scope of event extraction resources and methods to fully capture the semantics of statements relevant to biomolecular pathways.
Supporting Tools
- Argo - online environment for collaborative construction of text mining workflows and text annotation.
- brat - online environment for collaborative text annotation.
BioNLP 2013 Shared Task
To encourage the development of event extraction technology capable of pathway model curation support tasks, we are organizing the Pathway Curation event extraction task as part of the upcoming BioNLP Shared Task 2013.
We will provide task participants with documents relevant to reactions in a variety of signaling and metabolic pathways and full manual event annotation for these documents for use in the training and evaluation of event extraction methods. Please see the BioNLP Shared task 2013 page for more information and updates.
Project Team
NaCTeM Principal Investigator: Prof. Sophia AnaniadouNaCTeM researchers: Dr. Tomoko Ohta, Dr. Sampo Pyysalo, Dr. Makoto Miwa, Dr. Rafal Rak
NaCTeM software engineer: Dr. Andrew Rowley
KISTI Principal Investigator: Dr. Sung-Pil Choi
KISTI researcher: Dr. Hong-woo Chun
References
The following studies are relevant to the project:
- Brian Kemper, Takuya Matsuzaki Yukiko Matsuoka, Yoshimasa Tsuruoka, Hiroaki Kitano, Sophia Ananiadou and Jun'ichi Tsujii, PathText: a text mining integrator for biological pathway visualizations. Bioinformatics (2010) 26 (12): i374-i381.
- Tomoko Ohta, Sampo Pyysalo, Sophia Ananiadou and Jun'ichi Tsujii, Pathway Curation Support as an Information Extraction Task. In Proceedings of LBM 2011
- Tomoko Ohta, Sampo Pyysalo and Jun'ichi Tsujii, From Pathways to Biomolecular Events: Opportunities and Challenges. In Proceedings of BioNLP 2011.
- Rafal Rak, BalaKrishna Kolluru and Sophia Ananiadou. Building trainable taggers in a web-based, UIMA-supported NLP workbench. In Proceedings of ACL 2012 (To appear)
- Rafal Rak, Andrew Rowley and Sophia Ananiadou. Collaborative Development and Evaluation of Text-processing Workflows in a UIMA-supported Web-based Workbench. In Proceedings of LREC 2012, pp. 2971-2976
- Rafal Rak, Andrew Rowley, William J. Black, and Sophia Ananiadou. Argo: an integrative, interactive, text mining-based workbench supporting curation. Database: The Journal of Biological Databases and Curation, 2012.
- Pontus Stenetorp, Sampo Pyysalo, S., Goran Topić, Tomoko Ohta, Sophia Ananiadou, and Jun'ichi Tsujii. brat: a Web-based Tool for NLP-Assisted Text Annotation. In Proceedings of the Demonstrations at EACL, pp. 102-107.
Featured News
- 1st Workshop on Misinformation Detection in the Era of LLMs - Presentation slides now available
- Prof. Ananiadou appointed Deputy Director of the Christabel Pankhurst Institute
- ELLIS Workshop on Misinformation Detection - Presentation slides now available
- Prof. Sophia Ananiadou accepted as an ELLIS fellow
- BioNLP 2025 and Shared Tasks accepted for co-location at ACL 2025
- Prof. Junichi Tsujii honoured as Person of Cultural Merit in Japan
Other News & Events
- AI for Research: How Can AI Disrupt the Research Process?
- CL4Health @ NAACL 2025 - Extended submission deadline - 04/02/2025
- Invited talk at the 15th Marbach Castle Drug-Drug Interaction Workshop
- Participation in panel at Cyber Greece 2024 Conference, Athens
- Shared Task on Financial Misinformation Detection at FinNLP-FNP-LLMFinLegal








