EventMine - Event Extraction System for Biomedical Text
System Overview
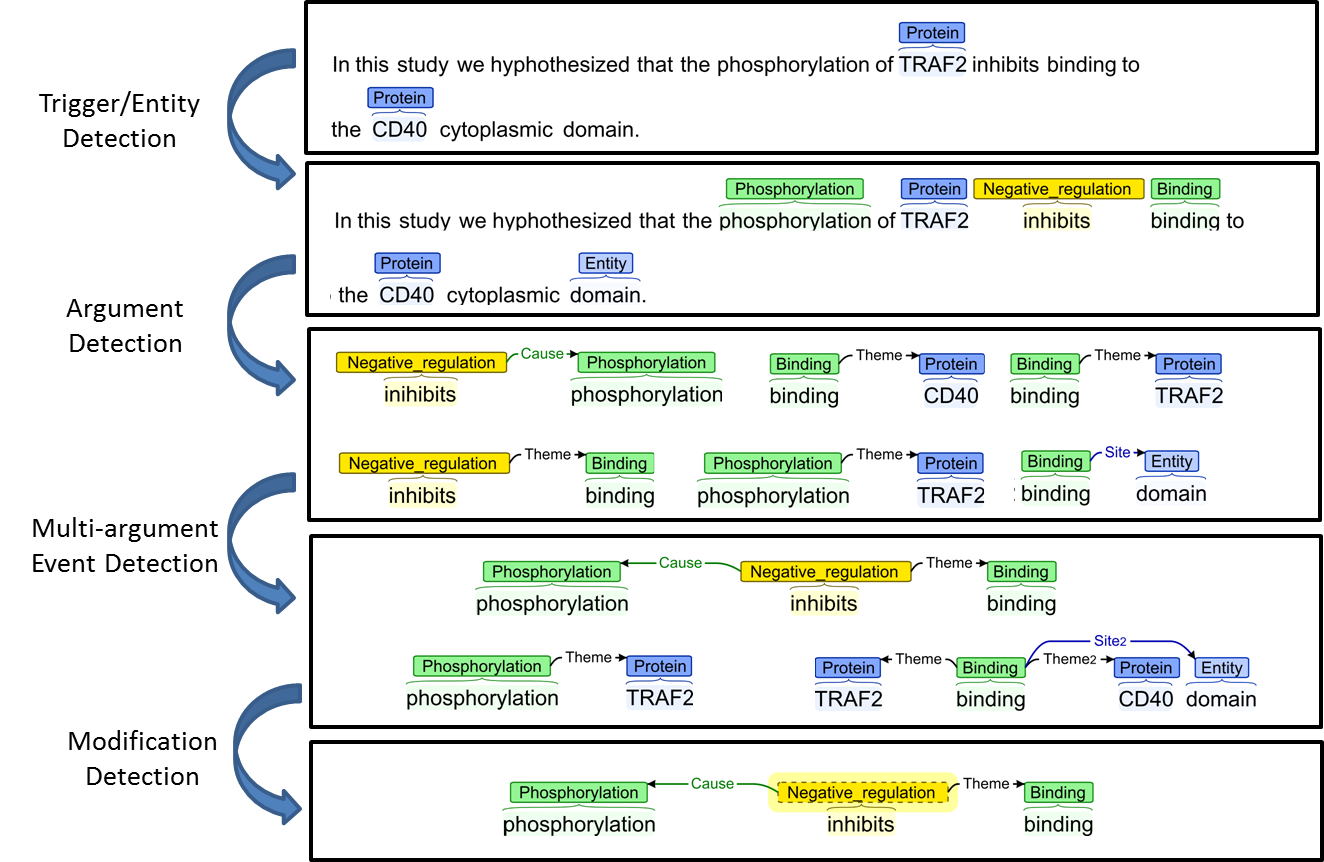
EventMine is a machine learning-based pipeline system, which extracts events from documents that already contain named entity annotations (e.g., genes/proteins, etc.). Given appropriate training data, it can be trained to extract many different types and structures of events. The core system consists of 4 detection modules, which operate on the output of syntactic parsers. These different modules are illustrated in Figure 1.
The modules illustrated are as follows:
- Trigger/Entity Detection - this module determines which words and phrases in a sentence potentially constitute participants of an event, and assigns appropriate types to them. These may be either entities, which play a role in the event (e.g., domain), or triggers, which are the words that characterise the type of the event, and around which the event is organised. In the example sentence shown, the words phospholylation, inhibits and binding are identified as potential event triggers for Phosphorylation, Negative regulation and Binding events
- Argument Detection - this module finds pairwise relations between event triggers and arguments, and assigns appropriate semantic types to the relations. In the example in Figure 1, six such relations have been found. The relations of type cause identify what is responsible for the event occurring. So, the event denoted by the the trigger phospholylation is responsible for the event denoted by inhibits. The relations of type theme denote entities or events that are affected by their parent event. So, for example, the Binding event is affected by the Negative regulation event, while TRAF2 is one of the entities affected by the Binding event. The relation of type site refers to a location where an event took place. In the case of figure 1, it refers to where the Binding event took place.
- Multi-argument Event Detection - this module combines individual pairwise relations into complete event structures. In the example shown in Figure 1, the six individual relations identified by the Argument Detection module have been grouped into 3 different event structures, with 1, 2 or 3 different arguments.
- Modification Detection - this module assigns modification information (i.e., negation and speculation) to each event. In the case of Figure 1, the presence of the word hypothesized means that the Negative regulation event is identified as being speculated.
In the BioNLP'09 Task 2 (i.e., the identification of secondary arguments, such as location and site), EventMine was able to outperform other systems that participated in this task.
Recent Developments
Recently, several modifications have been made to EventMine, both to improve its performance and its applicability to different tasks. The new version of EventMine was able to outperform other published systems on all tasks in the BioNLP'09 Task, GENIA, ID and COREF tasks in the BioNLP'11 Task.
- Coreference resolution - a new coreference system was developed and incorporated into EventMine. In order to correctly interpret certain events, coreference resolution is required. This is illustrated in Figure 2, in which there are two coreferential links. The first
link involves the mention this gene and its antecedent jun-B, while the second concerns the mention that and its antecedent expression. There are two gene expression events in the sentence, i.e., jun-B expression and jun-C expression, which can
only be correctly recognised if these coreferential links are identified and resolved.
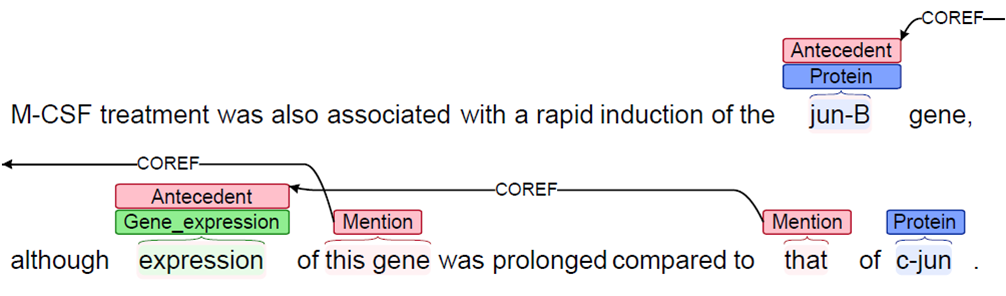 Figure 2. Event coreference example
Figure 2. Event coreference example - Domain Adaptation - The usual method of training and event extraction system is on a single annotated corpus. However, this can lead to problems of coverage and scalability. The new version of EventMine incorporates domain adaptation techniques, which aim to address this problem by allowing the use information from multiple annotated corpora when training the system.
-
Meta-knowledge identification - although the original EventMine has a Modification Detection module, there are still several other types of useful information that are present within the textual context of an event, and which can aid in its correct interpretation. This information includes whether an event represents a fact, hypothesis, experimental result or analysis of results, whether it describes new or previously reported knowledge and what is the intensity of the biological process described by the event. Together with information about negation and speculation, we refer to this interpretative information about events as meta-knowledge.
To better illustrate meta-knowledge, consider the following sentences. In each sentence, the event (triggered by the verb activate) and its participants (narL gene product as the CAUSE and nitrate reductase operon as the THEME) are identical. Hence, most event extraction systems, including the original EventMine, would not differentiate between the events. However, the textual context of the events means that the interpretation of the event is different in each case.
1. It is known that the narL gene product activates the nitrate reductase operon
2. We examined whether the narL gene product activates the nitrate reductase operon
3. The narL gene product partially activated the nitrate reductase operonIn sentence 1), the word known tells us that the event is a generally accepted fact, while in 2), the interpretation is completely different. The word examined denotes that the event is under investigation, and hence the truth value of the event is unknown. The word partially in sentence 3) does not challenge the truth of the event, but rather conveys the information that the strength or intensity of the event is less than may be expected by default.
EventMine has been enhanced so as to extract both events and five types of meta-knowledge information (i.e., Knowledge Type, Certainly Level, Manner, Polarity and Source) from biomedical literature. The automatically assigned meta-knowledge information can be used to refine search systems, which in turn can assist in several important tasks, e.g., database curation (by locating new experimental knowledge) and pathway enrichment (by providing information for inference). The meta-knowledge assignment system incorporated into EventMine has been trained on a version of the GENIA event corpus, whose 36,858 events have been manually enriched with meta-knowledge. Please see our meta-knowledge annotation page for further details.
In order to recognise meta-knowledge information, the EventMine pipeline has been slightly updated, so that the Trigger/Entity Detection module is now also able to recognise potential lexical cues (such as known, examined, etc.) that can be used to identify different types of meta-knowledge. Additionally, a Meta-Knowledge Assignment module, that assigns meta-knowledge values along the 5 different dimensions to each event, has also been incorporated into the pipeline.
References
Makoto Miwa, Rune Sætre, Jin-Dong Kim, and Jun'ichi Tsujii (2010). Event Extraction with Complex Event Classification Using Rich Features. Journal of Bioinformatics and Computational Biology (JBCB), 8(1): 131-146, February 2010.Makoto Miwa, Sampo Pyysalo, Tadayoshi Hara and Jun'ichi Tsujii (2010). A Comparative Study of Syntactic Parsers for Event Extraction. In Proceedings of the 2010 Workshop on Biomedical Natural Language Processing (BioNLP 2010), pp. 37-45.
Makoto Miwa, Sampo Pyysalo, Tadayoshi Hara and Jun'ichi Tsujii (2010). Evaluating Dependency Representation for Event Extraction. In Proceedings of the 23rd International Conference on Computational Linguistics (COLING 2010), pp. 779-787
Makoto Miwa, Paul Thompson and Sophia Ananiadou (2012). Boosting automatic event extraction from the literature using domain adaptation and coreference resolution. Bioinformatics
Makoto Miwa, Paul Thompson, John McNaught, Douglas B Kell and Sophia Ananiadou (2012). Extracting semantically enriched events from biomedical literature. BMC Bioinformatics, 13:108 (Highly Accessed)
Makoto Miwa, Sampo Pyysalo, Tomoko Ohta and Sophia Ananiadou (2013). Wide coverage biomedical event extraction using multiple partially overlapping corpora. BMC Bioinformatics, 14:175